Flipping and flopping without floundering
Changing my data from wide to long format, or vice versa was somehow always a headache. Eventhough I did this a lot, it just DID NOT stick in my head for the longest time. Everytime, I had to google it, and then work through at least 5 failed attempts before I’d get it. Therefore, this totally warrants a blog post, ya?
I recently started using tidyr’s spread() and gather() functions to do this, and I find them a lot more intuitive than the alternatives (the tidyverse strikes again), so that’s what I’ll focus on here. Other options, would be the reshape2 package, but honestly, I haven’t found any clear advantages of it over tidyr’s version, so I’ll ignore it for now…
Set up for some flippity floppity
So, let’s start by making some data that is in wide format. Let’s imagine we have 10 focal orangutans. Each orangutan has been followed to night-nest many times. Night nests can be built in one of 4 positions. So, each orangutan has a count of the number of times it has built a nest in each position. We’ll make the data in wide format with one column for the orangutan names, one column for the orangutan age-sex class, and one column for each nest position, where the counts will be held…
library(tidyverse)
## Warning: package 'ggplot2' was built under R version 3.5.2
## Warning: package 'tibble' was built under R version 3.5.2
## Warning: package 'dplyr' was built under R version 3.5.2
## Warning: package 'stringr' was built under R version 3.5.2
set.seed(100)
#generate the 6 columns
data <- as_tibble(cbind(Individual = c("Pi", "Roger", "Luaqlas", "Englas", "Boy", "Mindy", "Milo", "Loki", "Monster", "Jip"),
ASclass = c("AdultFemale", "FlangedMale", "AdolescentFemale", "AdolescentMale", "AdolescentMale",
"AdultFemale", "AdolescentFemale", "FlangedMale", "FlangedMale", "UnflangedMale"),
NestPos1 = sample(1:20, 10, replace = TRUE),
NestPos2 = sample(1:20, 10, replace = TRUE),
NestPos3 = sample(1:20, 10, replace = TRUE),
NestPos4 = sample(1:20, 10, replace = TRUE)))
#check it
str(data)
## Classes 'tbl_df', 'tbl' and 'data.frame': 10 obs. of 6 variables:
## $ Individual: chr "Pi" "Roger" "Luaqlas" "Englas" ...
## $ ASclass : chr "AdultFemale" "FlangedMale" "AdolescentFemale" "AdolescentMale" ...
## $ NestPos1 : chr "7" "6" "12" "2" ...
## $ NestPos2 : chr "13" "18" "6" "8" ...
## $ NestPos3 : chr "11" "15" "11" "15" ...
## $ NestPos4 : chr "10" "19" "7" "20" ...
#fix the numeric columns
data[,3:6] <- lapply(data[,3:6], as.numeric)
#check it all
data
## # A tibble: 10 x 6
## Individual ASclass NestPos1 NestPos2 NestPos3 NestPos4
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Pi AdultFemale 7 13 11 10
## 2 Roger FlangedMale 6 18 15 19
## 3 Luaqlas AdolescentFemale 12 6 11 7
## 4 Englas AdolescentMale 2 8 15 20
## 5 Boy AdolescentMale 10 16 9 14
## 6 Mindy AdultFemale 10 14 4 18
## 7 Milo AdolescentFemale 17 5 16 4
## 8 Loki FlangedMale 8 8 18 13
## 9 Monster FlangedMale 11 8 11 20
## 10 Jip UnflangedMale 4 14 6 3
summary(data)
## Individual ASclass NestPos1 NestPos2
## Length:10 Length:10 Min. : 2.00 Min. : 5.0
## Class :character Class :character 1st Qu.: 6.25 1st Qu.: 8.0
## Mode :character Mode :character Median : 9.00 Median :10.5
## Mean : 8.70 Mean :11.0
## 3rd Qu.:10.75 3rd Qu.:14.0
## Max. :17.00 Max. :18.0
## NestPos3 NestPos4
## Min. : 4.0 Min. : 3.00
## 1st Qu.: 9.5 1st Qu.: 7.75
## Median :11.0 Median :13.50
## Mean :11.6 Mean :12.80
## 3rd Qu.:15.0 3rd Qu.:18.75
## Max. :18.0 Max. :20.00Gather and Flip
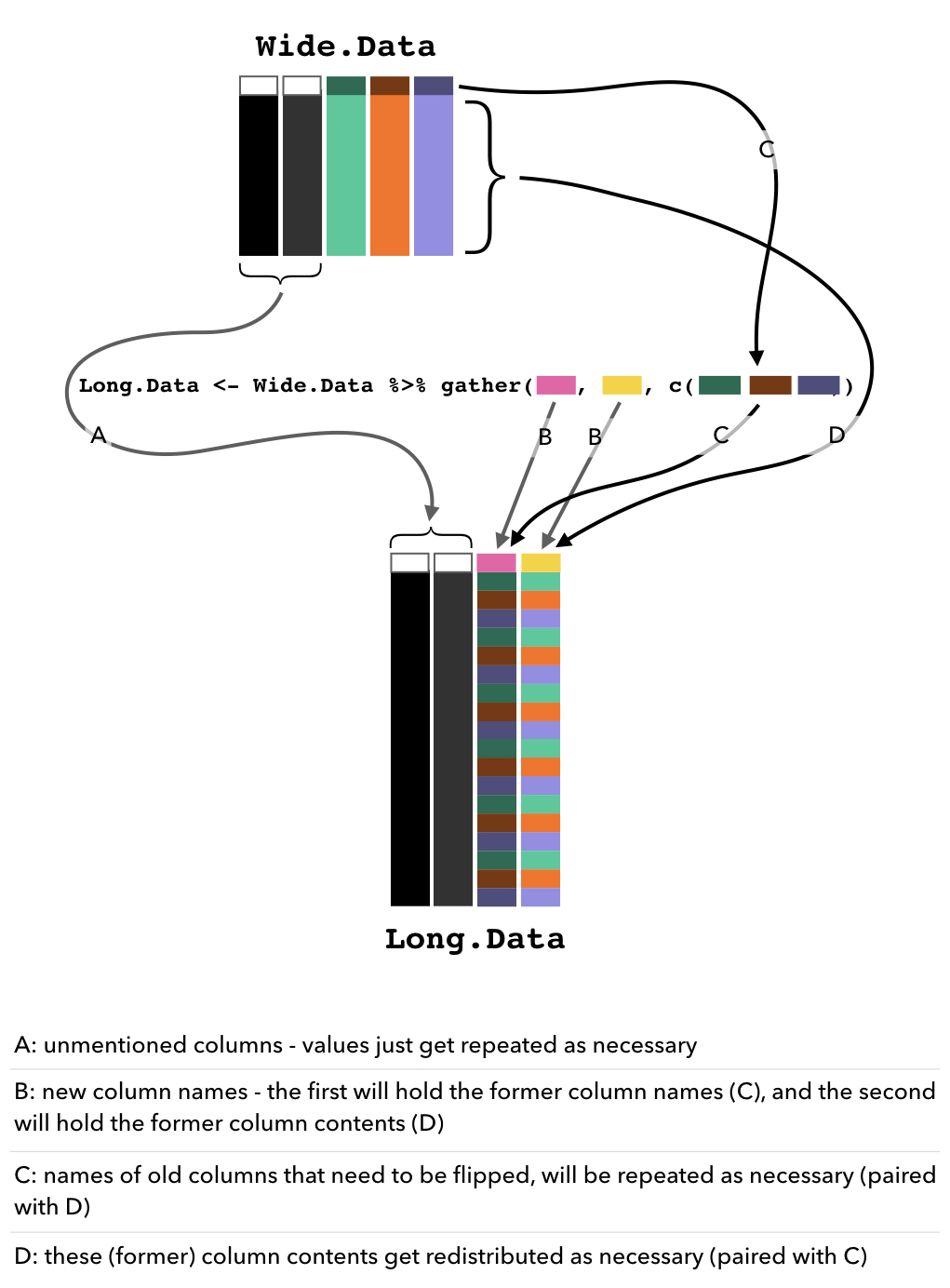
Ok, so now let’s say we want to FLIP the data down into LONG form - so we want one column for Individual, one column for ASclass, one column for NestPosition, and one column for the Counts of each nest position. As such, each Individual and ASclass pairing will be repeated FOUR times - once per nest position. The values in the NestPosition column should be 1, 2, 3, and 4, repeated for each Individual.
I will use the gather() function from the tidyr package…
#GATHER ((these two next lines of code are exactly the same))
data.long <- data %>% gather(NestPosition, Counts, 3:6) #use the column indices
data.long <- data %>% gather(NestPosition, Counts, c("NestPos1", "NestPos2", "NestPos3", "NestPos4")) #use the column names
#look at the whole thing
data.long %>% print(n = Inf)
## # A tibble: 40 x 4
## Individual ASclass NestPosition Counts
## <chr> <chr> <chr> <dbl>
## 1 Pi AdultFemale NestPos1 7
## 2 Roger FlangedMale NestPos1 6
## 3 Luaqlas AdolescentFemale NestPos1 12
## 4 Englas AdolescentMale NestPos1 2
## 5 Boy AdolescentMale NestPos1 10
## 6 Mindy AdultFemale NestPos1 10
## 7 Milo AdolescentFemale NestPos1 17
## 8 Loki FlangedMale NestPos1 8
## 9 Monster FlangedMale NestPos1 11
## 10 Jip UnflangedMale NestPos1 4
## 11 Pi AdultFemale NestPos2 13
## 12 Roger FlangedMale NestPos2 18
## 13 Luaqlas AdolescentFemale NestPos2 6
## 14 Englas AdolescentMale NestPos2 8
## 15 Boy AdolescentMale NestPos2 16
## 16 Mindy AdultFemale NestPos2 14
## 17 Milo AdolescentFemale NestPos2 5
## 18 Loki FlangedMale NestPos2 8
## 19 Monster FlangedMale NestPos2 8
## 20 Jip UnflangedMale NestPos2 14
## 21 Pi AdultFemale NestPos3 11
## 22 Roger FlangedMale NestPos3 15
## 23 Luaqlas AdolescentFemale NestPos3 11
## 24 Englas AdolescentMale NestPos3 15
## 25 Boy AdolescentMale NestPos3 9
## 26 Mindy AdultFemale NestPos3 4
## 27 Milo AdolescentFemale NestPos3 16
## 28 Loki FlangedMale NestPos3 18
## 29 Monster FlangedMale NestPos3 11
## 30 Jip UnflangedMale NestPos3 6
## 31 Pi AdultFemale NestPos4 10
## 32 Roger FlangedMale NestPos4 19
## 33 Luaqlas AdolescentFemale NestPos4 7
## 34 Englas AdolescentMale NestPos4 20
## 35 Boy AdolescentMale NestPos4 14
## 36 Mindy AdultFemale NestPos4 18
## 37 Milo AdolescentFemale NestPos4 4
## 38 Loki FlangedMale NestPos4 13
## 39 Monster FlangedMale NestPos4 20
## 40 Jip UnflangedMale NestPos4 3
str(data.long)
## Classes 'tbl_df', 'tbl' and 'data.frame': 40 obs. of 4 variables:
## $ Individual : chr "Pi" "Roger" "Luaqlas" "Englas" ...
## $ ASclass : chr "AdultFemale" "FlangedMale" "AdolescentFemale" "AdolescentMale" ...
## $ NestPosition: chr "NestPos1" "NestPos1" "NestPos1" "NestPos1" ...
## $ Counts : num 7 6 12 2 10 10 17 8 11 4 ...
summary(data.long)
## Individual ASclass NestPosition Counts
## Length:40 Length:40 Length:40 Min. : 2.00
## Class :character Class :character Class :character 1st Qu.: 7.00
## Mode :character Mode :character Mode :character Median :11.00
## Mean :11.03
## 3rd Qu.:15.00
## Max. :20.00It’s actually so simple that it surprises me every time. Let me try to break this down:
- I piped (
%>%) the original, wide form, data into thegatherfunction. - I gave it the new column names that I want: one name which will be the new column where the old column names are stored and repeated as necessary, and one name which will be the new column where all of the values from the old columns will be stored.
- I give it the indices of the columns that it should be rearranging.
Any unmentioned columns (in this example, Individual and ASclass) will keep their column names and the values within will just get repeated as necessary.
A desperate (and quite possibly, useless) attempt to represent this graphically:
Spread and Flop
Ok, so now let’s say we want to FLOP the data up into WIDE form - so, basically we want to do the opposite of what she just did above. We want to go from having one column per variable, to having only one row per orangutan - so we’d have one column for Individual, one column for ASclass, one column for each nest position (NestPos1 through to NestPos4), and the counts of each nest position will be in the respective columns.
I will use the spread() function from the tidyr package…
#SPREAD
data.wide <- data.long %>% spread(NestPosition, Counts)
#look at the whole thing
data.wide %>% print(n = Inf)
## # A tibble: 10 x 6
## Individual ASclass NestPos1 NestPos2 NestPos3 NestPos4
## <chr> <chr> <dbl> <dbl> <dbl> <dbl>
## 1 Boy AdolescentMale 10 16 9 14
## 2 Englas AdolescentMale 2 8 15 20
## 3 Jip UnflangedMale 4 14 6 3
## 4 Loki FlangedMale 8 8 18 13
## 5 Luaqlas AdolescentFemale 12 6 11 7
## 6 Milo AdolescentFemale 17 5 16 4
## 7 Mindy AdultFemale 10 14 4 18
## 8 Monster FlangedMale 11 8 11 20
## 9 Pi AdultFemale 7 13 11 10
## 10 Roger FlangedMale 6 18 15 19
str(data.wide)
## Classes 'tbl_df', 'tbl' and 'data.frame': 10 obs. of 6 variables:
## $ Individual: chr "Boy" "Englas" "Jip" "Loki" ...
## $ ASclass : chr "AdolescentMale" "AdolescentMale" "UnflangedMale" "FlangedMale" ...
## $ NestPos1 : num 10 2 4 8 12 17 10 11 7 6
## $ NestPos2 : num 16 8 14 8 6 5 14 8 13 18
## $ NestPos3 : num 9 15 6 18 11 16 4 11 11 15
## $ NestPos4 : num 14 20 3 13 7 4 18 20 10 19
summary(data.wide)
## Individual ASclass NestPos1 NestPos2
## Length:10 Length:10 Min. : 2.00 Min. : 5.0
## Class :character Class :character 1st Qu.: 6.25 1st Qu.: 8.0
## Mode :character Mode :character Median : 9.00 Median :10.5
## Mean : 8.70 Mean :11.0
## 3rd Qu.:10.75 3rd Qu.:14.0
## Max. :17.00 Max. :18.0
## NestPos3 NestPos4
## Min. : 4.0 Min. : 3.00
## 1st Qu.: 9.5 1st Qu.: 7.75
## Median :11.0 Median :13.50
## Mean :11.6 Mean :12.80
## 3rd Qu.:15.0 3rd Qu.:18.75
## Max. :18.0 Max. :20.00Again, it’s ridiculously simple:
- I piped (
%>%) the long form data into thespreadfunction. - I give it the name of the two columns that I want to be reorganized: the values from the first column will become the column names, and the values in the second column will be reorganized into those columns accordingly.
Any unmentioned columns (in this example, Individual and ASclass) will keep their column names and the values within will just get unduplicated as necessary.
That’s it. Easy Peasy once you get it to work. Inexplicably mind-boggling until then.